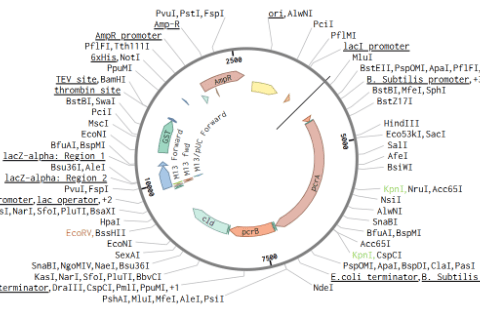
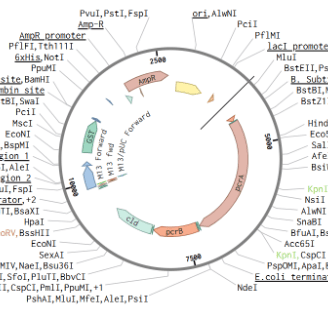
Our first plasmid design focused on reconstructing the perchlorate reduction pathway by incorporating the catalytic subunits of perchlorate reductase (pcrA and pcrB) alongside chlorite dismutase (Cld). Perchlorate reductase is a molybdenum-containing enzyme responsible for reducing perchlorate (ClO₄⁻) to chlorite (ClO₂⁻), while Cld further converts chlorite into chloride (Cl⁻) and molecular oxygen (O₂). This design served as more of an opportunity to learn how to properly use Benchling, to ensure the functionality of this design in the lab, we had to make refinements to the design.
Methods
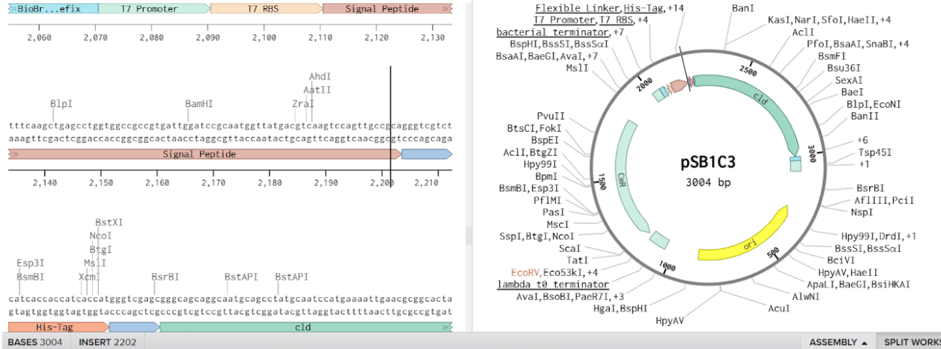
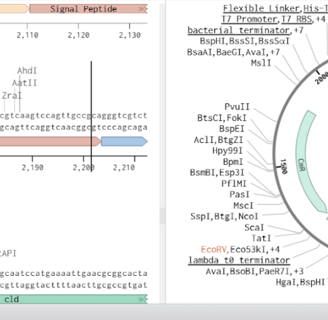
Our second plasmid design focused on expressing chlorite dismutase (Cld) alone for in-vitro characterization and structural analysis. To achieve high-yield expression, we used the pSB1C3 plasmid backbone, which supports a high copy number, ensuring robust gene expression in E. coli. The T7 promoter was chosen to drive strong, inducible expression, allowing precise control over protein production using IPTG induction. A periplasmic localization signal peptide was fused to the N-terminus of Cld to facilitate its transport into the periplasm, where disulfide bond formation enhances proper enzyme folding. Additionally, a 6x-Histidine tag (His-Tag) was added to the C-terminus of Cld, enabling efficient purification via Ni-NTA affinity chromatography. This construct was designed to facilitate high-throughput purification and biochemical characterization, allowing us to study enzyme kinetics, stability, and structural properties in detail. By optimizing expression and purification conditions, this plasmid supports further computational and experimental analysis of Cld activity, ultimately informing enzyme engineering strategies for improved bioremediation and oxygen production applications
Design 1: Perchlorate Reduction Pathway Construction
Design 2: Expression and Purification of Chlorite Dismutase
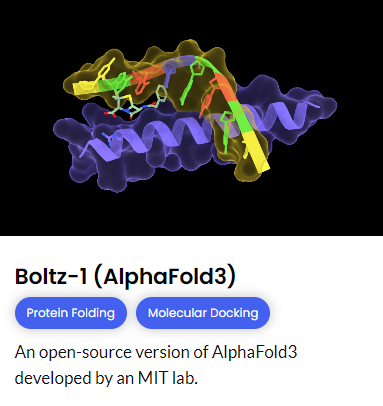
To investigate structural differences in Cld variants, we used RosettaTTFold2, Chai-1 (AlphaFold3), and Boltz-1 (AlphaFold3) to predict protein folding. This allowed us to visualize how specific mutations impact enzyme conformation and stability. Comparing our predicted models with Cld structures from previous studies, allowed us to indentify key structural features that may enhance enzyme efficiency.

Structural Modeling
Stability Analysis with Dynamut
To further analyze the effect of mutations on Cld stability, we used Dynamut, a computational tool that predicts changes in protein flexibility and stability based on single-point mutations. This helped us identify mutations that could improve enzyme robustness, potentially increasing its efficiency in perchlorate breakdown